Tidyverse, data manipulatione
Mikhail Dozmorov
Virginia Commonwealth University
10-13-2020
Tidyverse
The tidyverse is a collection of packages based on 4 principles for handling data:
- Reuse existing data structures
- Compose simple functions with the pipe
- Embrace functional programming
- Design for humans
The R project for Statistical Computing was built for a different age; the tidyverse is a collection of tools for our age
Reading in data
Base R functions for read-write the data
scan()- Read data into a vector or list from the console or fileread.table(),read.csv(),read.delim()- Reads a file in table format and creates a data frame from it, with cases corresponding to lines and variables to fields in the filewrite.table(),write.csv()- Saves the object (data.frame) to a file?data.table::freadfor very fast data read into R"File -> Import Dataset" in RStudio
readr
There are some built-in functions for reading in data in text files. These functions are read-dot-something, for example,
read.csv()reads in comma-delimited text data;read.delim()reads in tab-delimited text, etc.readrpackage provides fast and intelligent data reading capabilities. Very similar looking functions, named read-underscore-something -- e.g.,read_csv()They're good at guessing the types of data in the columns, they don't do some of the other silly things that the base functions do
Play nicely with
dplyr- data manipulation package
tibbles
Data frames are great! Except for
- printing them
- working with both characters and factors
- manipulating multiple columns
- You need to remember to set
options(stringsAsFactors = FALSE) - If you want a one-collumn data frame, you need to use
dat[, "column1", drop = FALSE]
tibbles are the data frame alternative simplifying work with data frame-like objects
tibbles
A
tibble, ortbl_df, is a modern reimagining of thedata.frame, keeping what time has proven to be effective, and throwing out what is notTibbles are
data.framesthat are lazy and surly: they do less (i.e., they don't change variable names or types, and don't do partial matching) and complain more (e.g., when a variable does not exist)This forces you to confront problems earlier, typically leading to cleaner, more expressive code. Tibbles also have an enhanced
printmethod which makes them easier to use with large datasets containing complex objects- Hadley Wickham, Chief Scientist at RStudio
glimpse()into tibble, analog ofstr()
Making the data tidy with tidyr
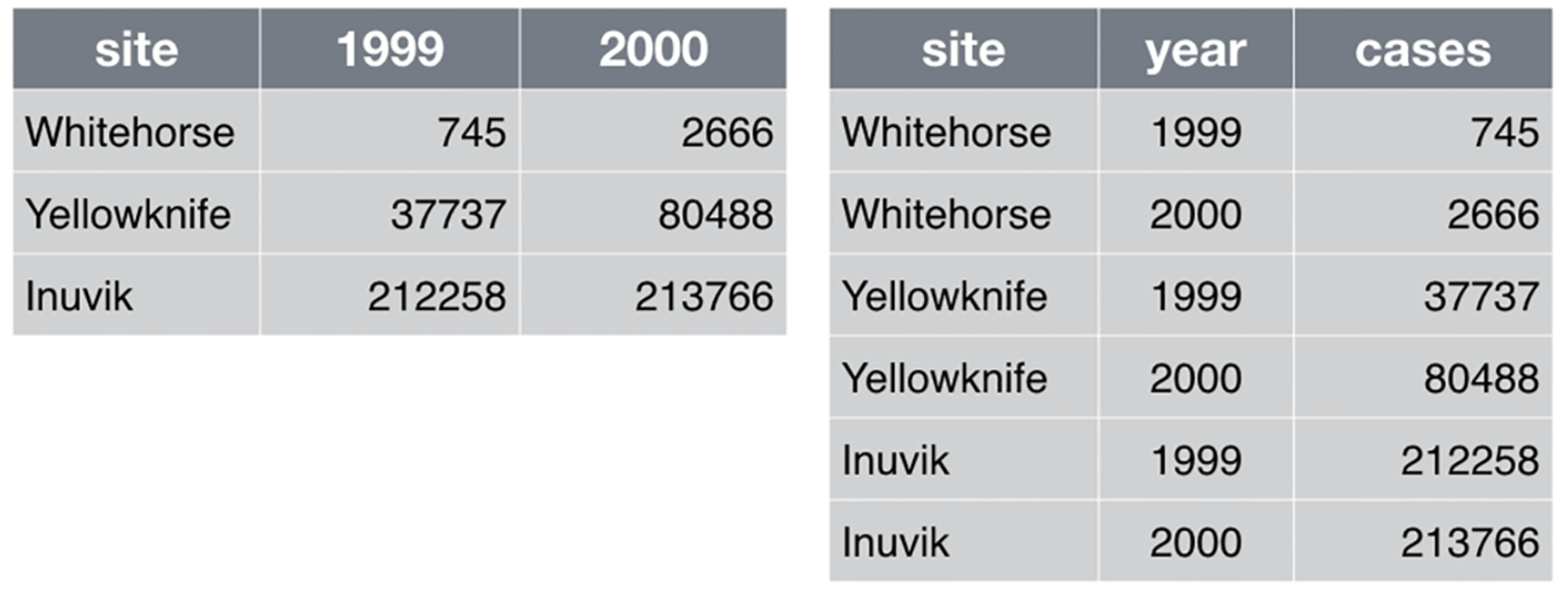
- Principles of tidy data
- Each column is a variable
- Each row is an observation
Tidy data paper, http://www.jstatsoft.org/v59/i10/paper
Making the data tidy with tidyr
tidyr- flexible data reshapingpivot_longer()- "lengthens" data, increasing the number of rows and decreasing the number of columnspivot_wider()- "widens" data, increasing the number of columns and decreasing the number of rows
Example of converting the wide data into tidy data

https://tidyr.tidyverse.org/index.html, vignette("tidy-data"), vignette("pivot")
Data manipulation with dplyr
dplyr: data manipulation with R
80% of your work will be data preparation
getting data (from databases, spreadsheets, flat-files)
performing exploratory/diagnostic data analysis
reshaping data
visualizing data
dplyr: data manipulation with R
80% of your work will be data preparation
Filtering rows (to create a subset)
Selecting columns of data (i.e., selecting variables)
Adding new variables
Sorting
Aggregating
Joining
dplyr: A grammar of data manipulation
The
dplyrpackage gives you a handful of useful verbs for managing data. On their own they don't do anything that base R can't doBasic
dplyrverbsfilter()select()mutate()arrange()summarize()group_by()
They all take a data frame or tibble as their input for the first argument, and they all return a data frame or tibble as output
The pipe %>% operator
- Pipe
%>%output of one command into an input of another command - chain commands together. (Think about the "|" operator in Linux) - Imported from
magrittrpackage - Read as "then". Take the dataset (or object), then do ...
library(dplyr)round( sqrt(1000), 3)## [1] 31.6231000 %>% sqrt %>% round(., 3)## [1] 31.623The pipe %>% operator
- For example, we can view the head of the
diamondsdata.frame using either of the last two lines of code here:
library(dplyr)library(ggplot2)data(diamonds)head(diamonds)diamonds %>% head## # A tibble: 6 x 10## carat cut color clarity depth table price x y z## <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43## 2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31## 3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31## 4 0.290 Premium I VS2 62.4 58 334 4.2 4.23 2.63## 5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75## 6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48The pipe %>% operator
For example, read the last line of code as: "Take the price column of the diamonds data.frame and then summarize it"
library(dplyr)data(diamonds)head(diamonds)diamonds %>% headsummary(diamonds$price)diamonds$price %>% summary(object = .)- There's a keyboard shortcut to insert the
%>%sequence - you can see what it is by clicking the Tools menu in RStudio, then selecting Keyboard Shortcut Help - On Mac, it's CMD-SHIFT-M
dplyr::filter()
If you want to filter rows of the data where some condition is true, use the filter() function.
- The first argument is the data frame you want to filter, e.g.
filter(mydata, .... - The second argument is a condition you must satisfy, e.g.
filter(ydat, symbol == "LEU1"). If you want to satisfy all of multiple conditions, you can use the "and" operator,&. The "or" operator|(the pipe character, usually shift-backslash) will return a subset that meet any of the conditions.
==: Equal to!=: Not equal to>,>=: Greater than, greater than or equal to<,<=: Less than, less than or equal to
dplyr::filter()
For example, keep only the entries with Ideal cut
df.diamonds_ideal <- filter(diamonds, cut == "Ideal")df.diamonds_ideal## # A tibble: 21,551 x 10## carat cut color clarity depth table price x y z## <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43## 2 0.23 Ideal J VS1 62.8 56 340 3.93 3.9 2.46## 3 0.31 Ideal J SI2 62.2 54 344 4.35 4.37 2.71## 4 0.3 Ideal I SI2 62 54 348 4.31 4.34 2.68## 5 0.33 Ideal I SI2 61.8 55 403 4.49 4.51 2.78## 6 0.33 Ideal I SI2 61.2 56 403 4.49 4.5 2.75## 7 0.33 Ideal J SI1 61.1 56 403 4.49 4.55 2.76## 8 0.23 Ideal G VS1 61.9 54 404 3.93 3.95 2.44## 9 0.32 Ideal I SI1 60.9 55 404 4.45 4.48 2.72## 10 0.3 Ideal I SI2 61 59 405 4.3 4.33 2.63## # … with 21,541 more rowsdplyr::filter()
We can achieve this same result using the %>% operator
diamonds %>% headdf.diamonds_ideal <- filter(diamonds, cut == "Ideal")df.diamonds_ideal <- diamonds %>% filter(cut == "Ideal")dplyr::select()
- The
filter()function allows you to return only certain rows matching a condition. Theselect()function returns only certain columns. The first argument is the data, and subsequent arguments are the columns you want.- Syntax:
select(data, columns)
- Syntax:
df.diamonds_ideal %>% headselect(df.diamonds_ideal, carat, cut, color, price, clarity)df.diamonds_ideal <- df.diamonds_ideal %>% select(., carat, cut, color, price, clarity)dplyr::mutate()
The
mutate()function adds new columns to the data that are functions of old columnsIt doesn't actually modify the data frame you're operating on, and the result is transient unless you assign it to a new object or reassign it back to itself (generally, not a good practice)
- Syntax:
mutate(data, new_column = function(old_columns))
- Syntax:
df.diamonds_ideal %>% headmutate(df.diamonds_ideal, price_per_carat = price/carat)df.diamonds_ideal <- df.diamonds_ideal %>% mutate(price_per_carat = price/carat)dplyr::arrange()
The
arrange()function does what it sounds like - sorts thingsIt takes a
data.frameortbl_dfand arranges (or sorts) by column(s) of interestThe first argument is the data, and subsequent arguments are columns to sort on. Use the
desc()function to arrange by descending- Syntax:
arrange(data, column_to_sort_by)
- Syntax:
df.diamonds_ideal %>% headarrange(df.diamonds_ideal, price)df.diamonds_ideal %>% arrange(price, price_per_carat)dplyr::summarize()
The
summarize()function summarizes multiple values to a single valueThe power of
summarize()comes from a few convenience functions calledn()andn_distinct()that tell you the number of observations or the number of distinct values of a particular variable.- Syntax:
summarize(function_of_variables)
- Syntax:
summarize(df.diamonds_ideal, length = n(), avg_price = mean(price))df.diamonds_ideal %>% summarize(length = n(), avg_price = mean(price))dplyr::group_by()
Summarize subsets of columns by custom summary statistics
Syntax:
group_by(data, column_to_group)
group_by(diamonds, cut) %>% summarize(mean(price))group_by(diamonds, cut, color) %>% summarize(mean(price))The power of pipe %>%
- Summarize subsets of columns by custom summary statistics
arrange(mutate(arrange(filter(tbl_df(diamonds), cut == "Ideal"), price), price_per_carat = price/carat), price_per_carat)arrange( mutate( arrange( filter(tbl_df(diamonds), cut == "Ideal"), price), price_per_carat = price/carat), price_per_carat)diamonds %>% filter(cut == "Ideal") %>% arrange(price) %>% mutate(price_per_carat = price/carat) %>% arrange(price_per_carat)Joining data frames
inner_join(x, y): Keep only rows where there are observations in bothxandyleft_join(x, y): Keep all rows fromx, whether they have a match inyor not (unmatched rows are filled with NAs)right_join(x, y): Keep all rows fromy, whether they have a match inxor notfull_join(x, y): Keep all rows from bothxandy, whether they have a match in the other dataset or not
Review https://ready4r.netlify.app/labbook/part-5-doing-useful-things-with-multiple-tables.html#joining-tables
Working with factors tidyverse way
library(forcats)
fct_rev()- Reverse order of factor levelsfct_reorder()- Reordering a factor by another variablefct_collapse()- Collapse multiple categories into one categoryfct_lump()- Collapsing the least/most frequent values of a factor into “other”fct_infreq()- Reordering a factor by the frequency of valuesfct_relevel()- Changing the order of a factor by hand
https://forcats.tidyverse.org/